Difference between revisions of "CSC334 lab7"
| Line 1: | Line 1: | ||
| − | + | =Introduction= | |
A good definition of sequence logos can be found in Wikipedia: | A good definition of sequence logos can be found in Wikipedia: | ||
| Line 5: | Line 5: | ||
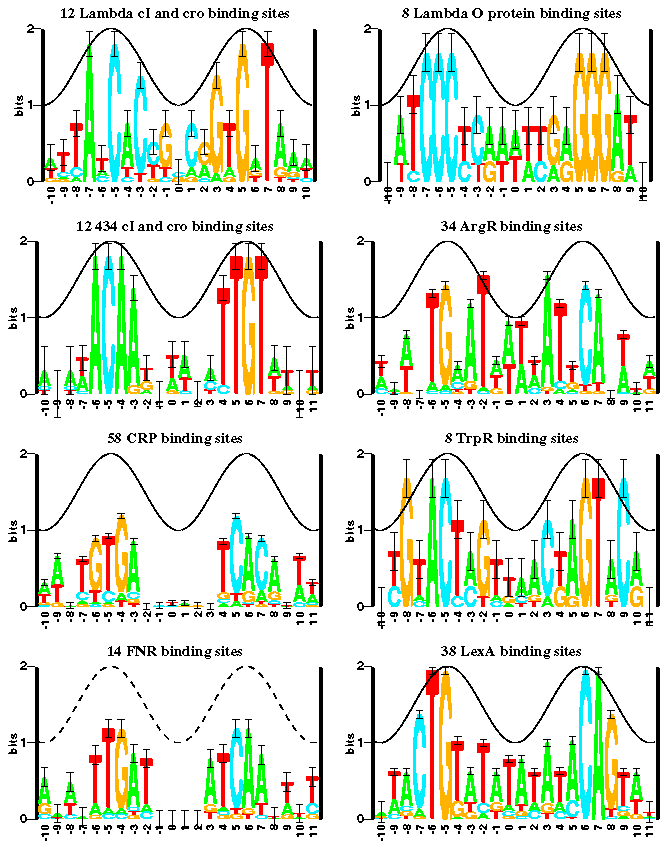
::To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. '''The sequence logo will show how well residues are conserved at each position: the fewer the number of residues, the higher the letters will be, because the better the conservation is at that position'''. Different residues at the same position will be scaled according to their frequency. Sequence logos can be used to represent conserved DNA binding sites, where transcription factors bind. [http://en.wikipedia.org/wiki/Sequence_logo] | ::To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. '''The sequence logo will show how well residues are conserved at each position: the fewer the number of residues, the higher the letters will be, because the better the conservation is at that position'''. Different residues at the same position will be scaled according to their frequency. Sequence logos can be used to represent conserved DNA binding sites, where transcription factors bind. [http://en.wikipedia.org/wiki/Sequence_logo] | ||
| + | |||
| + | [[Image:sequence_logo.png]] | ||
| + | This image is take from a the following document that you should read to get a good start on this lab: | ||
| + | [http://www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/ www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/] | ||
| + | |||
| + | =Lab= | ||
| + | |||
| + | ==The Sequences== | ||
For this lab we will use 8 different sequences: | For this lab we will use 8 different sequences: | ||
| Line 24: | Line 32: | ||
String seq[8]; | String seq[8]; | ||
</pre></code> | </pre></code> | ||
| + | |||
| + | ==First step: Define the window== | ||
| + | |||
| + | More information will be provided during the lab. The goal of this step is to define the geometry of the window and the constants used by the program. | ||
Revision as of 12:34, 4 August 2008
Contents
Introduction
A good definition of sequence logos can be found in Wikipedia:
- A sequence logo in bioinformatics is a graphical representation of the sequence conservation of nucleotides (in a strand of DNA/RNA) or amino acids (in protein sequences) [1]
- To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. The sequence logo will show how well residues are conserved at each position: the fewer the number of residues, the higher the letters will be, because the better the conservation is at that position. Different residues at the same position will be scaled according to their frequency. Sequence logos can be used to represent conserved DNA binding sites, where transcription factors bind. [2]
 This image is take from a the following document that you should read to get a good start on this lab:
www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/
This image is take from a the following document that you should read to get a good start on this lab:
www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/
Lab
The Sequences
For this lab we will use 8 different sequences:
seq[0] = "CCCATTGTTCTC";
seq[1] = "TTTCTGGTTCTC";
seq[2] = "TCAATTGTTTAG";
seq[3] = "CTCATTGTTGTC";
seq[4] = "TCCATTGTTCTC";
seq[5] = "CCTATTGTTCTC";
seq[6] = "TCCATTGTTCGT";
seq[7] = "CCAATTGTTTTG";
They are shown here as taken from a Processing program where the sequences are stored in an array of 8 strings:
String seq[8];
First step: Define the window
More information will be provided during the lab. The goal of this step is to define the geometry of the window and the constants used by the program.