Difference between revisions of "CSC334 Lab8"
(→Technical University of Denmark Logo Generator) |
|||
| Line 88: | Line 88: | ||
</pre></code> | </pre></code> | ||
| − | + | =Questins= | |
| + | # Why is the information reported larger than 2.0? | ||
| + | # On the input page of the Technical University of Denmark, you will notice a line above an input box: | ||
| + | <code><pre> | ||
| + | A priori amino acid distribution (4 decimal precision): | ||
| + | pA; pC; pD; pE; pF; pG; pH; pI; pK; pL; pM; pN; pP; pQ; pR; pS; pT; pV; pW; pY; | ||
| + | 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 | ||
| + | </pre></code> | ||
| + | |||
| + | ::What do the the 0.05 numbers refer to? | ||
Revision as of 18:07, 4 August 2008
<meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> <meta name="description" content="DNA Sequence Logo Lab" /> <meta name="title" content="Bioinformatics Lab" /> <meta name="abstract" content="DNA Sequence Logo" /> <meta name="author" content="thiebaut at cs.smith.edu" />
Contents
Introduction
This lab should be done in conjunction with Lab 7 dealing with DNA sequence logos. Please refer to the introduction section of Lab 7 for more information on sequence logos.
Play sequences
We are going to use the following sequences in this lab, but feel free to use your own sequences
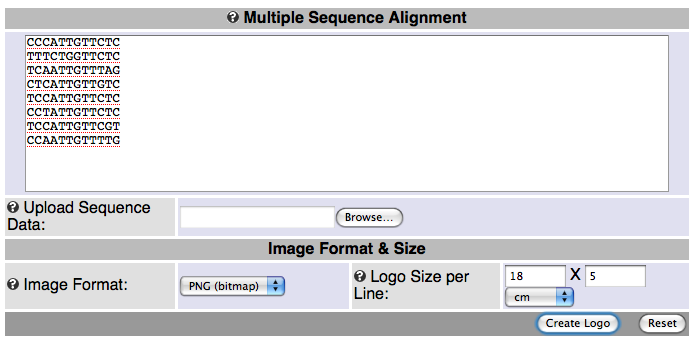
CCCATTGTTCTC TTTCTGGTTCTC TCAATTGTTTAG CTCATTGTTGTC TCCATTGTTCTC CCTATTGTTCTC TCCATTGTTCGT CCAATTGTTTTG
Logos on the Web
Berkeley Logo Generator
Point your browser to weblogo.berkeley.edu, which is one of the currently better logo generators on the Web.
Enter the 8 sequences of Lab 7 in the input window, and click on the Create button:
Compare the output to the one you obtained in Lab 7.
Technical University of Denmark Logo Generator
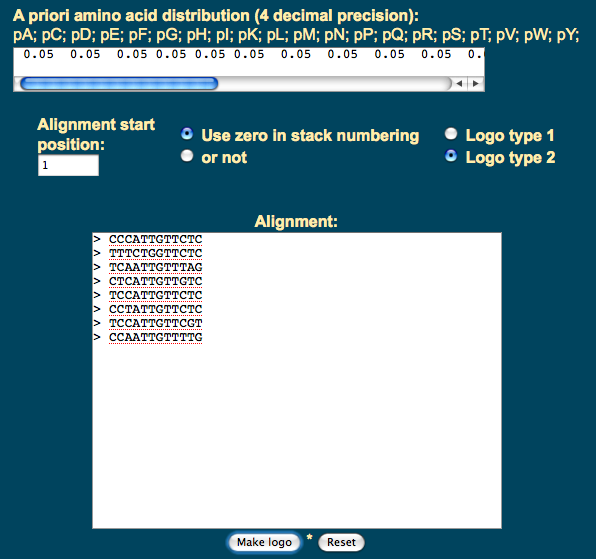
Another option is to point your browser to www.cbs.dtu.dk/~gorodkin/appl/plogo.html, and to enter the same eight sequences, as illustrated below:
Verify that you get the same logo as in the previous step.
You will notice that the logo output also gives the following results:
Your data:
> CCCATTGTTCTC
> TTTCTGGTTCTC
> TCAATTGTTTAG
> CTCATTGTTGTC
> TCCATTGTTCTC
> CCTATTGTTCTC
> TCCATTGTTCGT
> CCAATTGTTTTG
Your amino acid distribution:
0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05
The distribution of information over the alignment:
position information/entropy
1 3.3219
2 3.5106
3 2.8219
4 3.7784
5 4.3219
6 3.7784
7 4.3219
8 4.3219
9 4.3219
10 3.0231
11 3.2606
12 3.0231
Information content for the whole alignment: 43.8059
Questins
- Why is the information reported larger than 2.0?
- On the input page of the Technical University of Denmark, you will notice a line above an input box:
A priori amino acid distribution (4 decimal precision):
pA; pC; pD; pE; pF; pG; pH; pI; pK; pL; pM; pN; pP; pQ; pR; pS; pT; pV; pW; pY;
0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05 0.05
- What do the the 0.05 numbers refer to?