Difference between revisions of "CSC334 Lab4"
(→Solution Program) |
|||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> | ||
| + | <meta name="description" content="DNA Sequence Logo Lab" /> | ||
| + | <meta name="title" content="Bioinformatics Lab" /> | ||
| + | <meta name="abstract" content="DNA Sequence Logo" /> | ||
| + | <meta name="author" content="thiebaut at cs.smith.edu" /> | ||
| + | |||
| + | |||
| + | |||
[[Csc334_Labs | Back to CSC334 Lab Page]] | [[Csc334_Labs | Back to CSC334 Lab Page]] | ||
<hr /><br /> | <hr /><br /> | ||
| Line 5: | Line 13: | ||
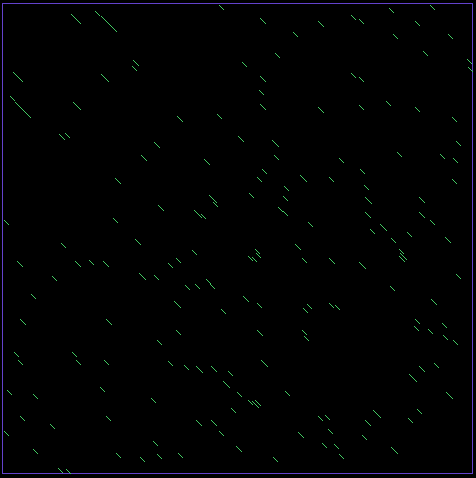
In this lab you will work with a Proce55ing sketchbook to create a graphical representation of repeating sequences in DNA sequences. Finding repeating patterns, either adjacent to each other, as in [http://en.wikipedia.org/wiki/Tandem_repeat tandom repeats], or long [http://en.wikipedia.org/wiki/Interspersed_repeat interspersed repeats] yield important genetic information about a DNA sequence. | In this lab you will work with a Proce55ing sketchbook to create a graphical representation of repeating sequences in DNA sequences. Finding repeating patterns, either adjacent to each other, as in [http://en.wikipedia.org/wiki/Tandem_repeat tandom repeats], or long [http://en.wikipedia.org/wiki/Interspersed_repeat interspersed repeats] yield important genetic information about a DNA sequence. | ||
| + | |||
| + | Repeats can also be found comparing the sequence to its reverse. Biologists are interested in repeats because they are often indicative of genome rearrangements. | ||
[[Image:findingDNARepeats.png]] | [[Image:findingDNARepeats.png]] | ||
| Line 13: | Line 23: | ||
* Create the font needed for the program. Click on '''Tools''', '''Create Font''', and select '''Monaco''', with a size of 12 points. | * Create the font needed for the program. Click on '''Tools''', '''Create Font''', and select '''Monaco''', with a size of 12 points. | ||
This will create the file ''Monaco-12.vlw'' in the data folder of your Processing ''sketchbook''. If you cannot find the font Monaco on your computer, select a [http://en.wikipedia.org/wiki/Typeface#Monospaced_typefaces monospace font] such as '''Prestige Elite''', or '''Courier'''. | This will create the file ''Monaco-12.vlw'' in the data folder of your Processing ''sketchbook''. If you cannot find the font Monaco on your computer, select a [http://en.wikipedia.org/wiki/Typeface#Monospaced_typefaces monospace font] such as '''Prestige Elite''', or '''Courier'''. | ||
| + | * Run the program. Verify that you get a empty square and a listing of the DNA sequence | ||
| + | ===Step 1: Finding repeats=== | ||
| + | This part involves the whole class to discover the algorithm that can be used to find repeat patterns longer than some predefined length. | ||
| + | * come up with an algorithm (this will take some time!) | ||
| + | * code it, and test it. Use a minimum length of 1 or 2 at first, and increase it slowly to see the longest pattern founds. | ||
| + | * verify that you get an image similar to the one above. | ||
| + | ===Step 2: finding the longest repeat === | ||
| + | * Make your program print in the console the locations of the longest repeats it finds, in the following form: | ||
| + | longest repeat: 123, 255 length 7 CGGTAAC | ||
| + | This means that a sequence of length 7 was found at Index 123 and at Index 255, and reads CGGTAAC | ||
| + | ===Step 3: Marking the longest repeat=== | ||
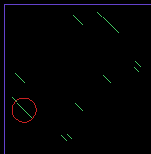
| + | * Once your program works, make it display a circle around the longest repeat (or repeats) found. Use the '''drawPoints( int i, int j, int n )''' function as an example for a new function '''putCircleAround( int i, int j, int radius )''' that could be used to highlight the longest segment. | ||
| + | [[Image:DNARepeats_circle.png | center ]] | ||
| + | Note that to draw a circle that is not filled with color, you use the '''noFill()''' function and call '''ellipse()'''. | ||
| − | + | noFill(); | |
| − | + | ellipse( x, y, radius, radius ); | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 18:13, 4 August 2008
<meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> <meta name="description" content="DNA Sequence Logo Lab" /> <meta name="title" content="Bioinformatics Lab" /> <meta name="abstract" content="DNA Sequence Logo" /> <meta name="author" content="thiebaut at cs.smith.edu" />
Contents
Finding repeats in DNA sequences
In this lab you will work with a Proce55ing sketchbook to create a graphical representation of repeating sequences in DNA sequences. Finding repeating patterns, either adjacent to each other, as in tandom repeats, or long interspersed repeats yield important genetic information about a DNA sequence.
Repeats can also be found comparing the sequence to its reverse. Biologists are interested in repeats because they are often indicative of genome rearrangements.
Methodology
- Copy and paste the program DNA_Repeats_start.pde in a new sketchbook which you should call DNA_Repeats.pde.
- Create the font needed for the program. Click on Tools, Create Font, and select Monaco, with a size of 12 points.
This will create the file Monaco-12.vlw in the data folder of your Processing sketchbook. If you cannot find the font Monaco on your computer, select a monospace font such as Prestige Elite, or Courier.
- Run the program. Verify that you get a empty square and a listing of the DNA sequence
Step 1: Finding repeats
This part involves the whole class to discover the algorithm that can be used to find repeat patterns longer than some predefined length.
- come up with an algorithm (this will take some time!)
- code it, and test it. Use a minimum length of 1 or 2 at first, and increase it slowly to see the longest pattern founds.
- verify that you get an image similar to the one above.
Step 2: finding the longest repeat
- Make your program print in the console the locations of the longest repeats it finds, in the following form:
longest repeat: 123, 255 length 7 CGGTAAC
This means that a sequence of length 7 was found at Index 123 and at Index 255, and reads CGGTAAC
Step 3: Marking the longest repeat
- Once your program works, make it display a circle around the longest repeat (or repeats) found. Use the drawPoints( int i, int j, int n ) function as an example for a new function putCircleAround( int i, int j, int radius ) that could be used to highlight the longest segment.
Note that to draw a circle that is not filled with color, you use the noFill() function and call ellipse().
noFill(); ellipse( x, y, radius, radius );
Solution Program
Back to CSC334 Lab Page
© D. Thiebaut 2008