Difference between revisions of "CSC334 Lab5"
(New page: Back to CSC334 Lab Page <hr /><br /> =Finding Repeats using a Web-Based Repeat Finder= In this lab we perform a similar operation as the one we did in CSC334_Lab4,...) |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | <meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> | ||
| + | <meta name="description" content="DNA Sequence Logo Lab" /> | ||
| + | <meta name="title" content="Bioinformatics Lab" /> | ||
| + | <meta name="abstract" content="DNA Sequence Logo" /> | ||
| + | <meta name="author" content="thiebaut at cs.smith.edu" /> | ||
| + | |||
| + | |||
[[Csc334_Labs | Back to CSC334 Lab Page]] | [[Csc334_Labs | Back to CSC334 Lab Page]] | ||
<hr /><br /> | <hr /><br /> | ||
| Line 7: | Line 14: | ||
Make sure you have a FASTA sequence available, and follow these steps | Make sure you have a FASTA sequence available, and follow these steps | ||
| − | * Point your browser to the [http://arbl.cvmbs.colostate.edu/molkit/ Molecular Toolkit] at Colorado State U. | + | * Point your browser to the [http://arbl.cvmbs.colostate.edu/molkit/ Molecular Toolkit] at Colorado State U. ([http://arbl.cvmbs.colostate.edu/molkit/]) |
| − | * Select the '''Dot Plot''' option | + | * Select the '''Dot Plot''' option and jump to the '''Nucleic Acid Dot Plots'''tom page. |
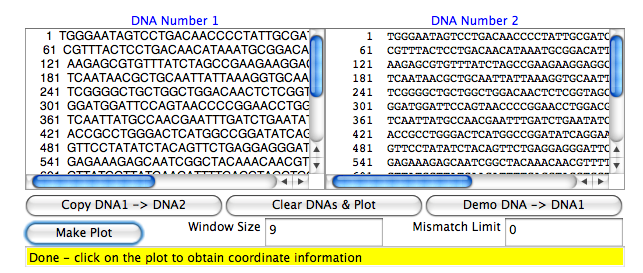
* Enter your favorite DNA sequence in the left text area, and click on '''Copy DNA1->DNA2''' | * Enter your favorite DNA sequence in the left text area, and click on '''Copy DNA1->DNA2''' | ||
[[Image:DNA_DotPlot.png]] | [[Image:DNA_DotPlot.png]] | ||
| + | * Select window sizes that are odd number, and generate different plots. Observe that you observe similar data as what you obtained in [[Csc334_Lab4 | Lab #4]] if you did this lab. | ||
| + | |||
| + | * Click on the segments in the dot plot, and observe that you can get some idea of the location of the repeating segment's locations in the DNA sequence. | ||
| + | |||
| + | [[Image:DNA_DotPlot2.png]] | ||
| + | |||
| + | * What about finding repeats between the sequence and its reverse version? Or between it and its complementary version? The Molecular Toolkit provides different tools for processing sequences. Go back to the Molecular Toolkit [http://arbl.cvmbs.colostate.edu/molkit/ home page] and click on '''Manipulate and Display a DNA Sequence''' | ||
| + | |||
| + | * Try the different options available, and see that you get different versions of your DNA sequence. This really shows how much a sequence is really one of a pair that can be read forward or backward! For the purpose of this exercise finally click on '''Inverse Complement''' and copy to your clipboard the sequence obtained. | ||
| + | |||
| + | * Paste it in the DNA1 window of the dot-plot tool, and paste your original sequence in the DNA2 window, and generate a new dot plot. | ||
| + | |||
| + | That's it for this lab. In combination with [[CSC334_Lab4]], this will have given you some exposure to the problem of finding repeats in a sequence. | ||
<br /> | <br /> | ||
Latest revision as of 18:13, 4 August 2008
<meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> <meta name="description" content="DNA Sequence Logo Lab" /> <meta name="title" content="Bioinformatics Lab" /> <meta name="abstract" content="DNA Sequence Logo" /> <meta name="author" content="thiebaut at cs.smith.edu" />
Finding Repeats using a Web-Based Repeat Finder
In this lab we perform a similar operation as the one we did in CSC334_Lab4, but this time using a Web service.
Make sure you have a FASTA sequence available, and follow these steps
- Point your browser to the Molecular Toolkit at Colorado State U. ([1])
- Select the Dot Plot option and jump to the Nucleic Acid Dot Plotstom page.
- Enter your favorite DNA sequence in the left text area, and click on Copy DNA1->DNA2
- Select window sizes that are odd number, and generate different plots. Observe that you observe similar data as what you obtained in Lab #4 if you did this lab.
- Click on the segments in the dot plot, and observe that you can get some idea of the location of the repeating segment's locations in the DNA sequence.
- What about finding repeats between the sequence and its reverse version? Or between it and its complementary version? The Molecular Toolkit provides different tools for processing sequences. Go back to the Molecular Toolkit home page and click on Manipulate and Display a DNA Sequence
- Try the different options available, and see that you get different versions of your DNA sequence. This really shows how much a sequence is really one of a pair that can be read forward or backward! For the purpose of this exercise finally click on Inverse Complement and copy to your clipboard the sequence obtained.
- Paste it in the DNA1 window of the dot-plot tool, and paste your original sequence in the DNA2 window, and generate a new dot plot.
That's it for this lab. In combination with CSC334_Lab4, this will have given you some exposure to the problem of finding repeats in a sequence.
Back to CSC334 Lab Page