Difference between revisions of "Csc334 DT lab Ideas"
| (16 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<hr> | <hr> | ||
| − | * Get a DNA sequence [[CSC334_Lab1 Lab #1]] | + | * Get familiar with Proce55ing [[CSC334_Lab0| Lab #0]] |
| + | * Get a DNA sequence [[CSC334_Lab1| Lab #1]] | ||
| + | * Get a protein sequence [[CSC334_Lab2 | Lab #2]] | ||
* given a series of nucleotides, find and display the 6 reading frames | * given a series of nucleotides, find and display the 6 reading frames | ||
* finding repeats within a sequence | * finding repeats within a sequence | ||
* BLAST lab: take a sequence (protein and/or DNA) and find other sequences that are close to it | * BLAST lab: take a sequence (protein and/or DNA) and find other sequences that are close to it | ||
* given N sequences that overlap, find the best way in which they form a long sequence. Use processing and a circle approach to represent them | * given N sequences that overlap, find the best way in which they form a long sequence. Use processing and a circle approach to represent them | ||
| − | * display alignment of two sequences | + | * display alignment of two sequences [[CSC334_Lab3| Lab #3]] |
** limit to end to end overlapping | ** limit to end to end overlapping | ||
** from output of blast | ** from output of blast | ||
** from result of computing local alignment | ** from result of computing local alignment | ||
| + | |||
| + | * Orfing: (page 146) Finding a sequence longer than some number (proteins are typically 350 residues in length, and at least 100 minimum), between a start (ATG), and a stop (TAA, TAG, TGA) sequence. | ||
| + | |||
| + | * [[Image:FoldIt.png | left | thumb | 100px ]] | ||
| + | <br /> | ||
| + | FoldIt! [http://fold.it/portal/adobe_main http://fold.it]. Read NYT Article | ||
| + | [http://www.nytimes.com/2008/07/22/science/22inno.html?pagewanted=all If You Have a Problem, Use Innocentive to Ask Everyone] for background. | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | |||
| + | * Could do a few labs from the NCBI 12 mini-course Web page. ([http://www.ncbi.nlm.nih.gov/Class/minicourses/ www.ncbi.nlm.nih.gov/Class/minicourses]) | ||
| + | |||
| + | ''I tried a few... they are fairly cryptic and require good knowledge of biological terms...'' | ||
| + | |||
| + | ** Bioinformatics Quick Start | ||
| + | ** Making Sense of DNA and Protein Sequences | ||
| + | ** Unmasking Genes in Human DNA | ||
| + | ** Identification of Disease Genes | ||
| + | **Correlating Disease Genes and Phenoypes | ||
| + | ** BLAST Quick Start | ||
| + | ** EntrezGene Quick Start | ||
| + | ** Structure Analysis Quick Start | ||
| + | ** MapViewer Quick Start | ||
| + | ** GenBank Quick Start | ||
| + | ** Entrez Quick Start | ||
| + | ** Microbial Genomes Quick Start | ||
| + | |||
| + | * Lab on Sequence Logo with Processing. Good reference can be found [http://rantamplan.bio.ub.es/~eblanco/courses/papers/dhaeseleer2006.pdf here]. | ||
| + | [[Image:sequenceLogoEcoli.png]] | ||
| + | <br /> | ||
| + | Image taken from [http://www.clcbio.com/index.php?id=869 http://www.clcbio.com/index.php?id=869] | ||
| + | <br /> | ||
<hr> | <hr> | ||
[[CSC334 | Back to CSC334]] | [[CSC334 | Back to CSC334]] | ||
Latest revision as of 11:03, 1 August 2008
- Get familiar with Proce55ing Lab #0
- Get a DNA sequence Lab #1
- Get a protein sequence Lab #2
- given a series of nucleotides, find and display the 6 reading frames
- finding repeats within a sequence
- BLAST lab: take a sequence (protein and/or DNA) and find other sequences that are close to it
- given N sequences that overlap, find the best way in which they form a long sequence. Use processing and a circle approach to represent them
- display alignment of two sequences Lab #3
- limit to end to end overlapping
- from output of blast
- from result of computing local alignment
- Orfing: (page 146) Finding a sequence longer than some number (proteins are typically 350 residues in length, and at least 100 minimum), between a start (ATG), and a stop (TAA, TAG, TGA) sequence.
FoldIt! http://fold.it. Read NYT Article
If You Have a Problem, Use Innocentive to Ask Everyone for background.
- Could do a few labs from the NCBI 12 mini-course Web page. (www.ncbi.nlm.nih.gov/Class/minicourses)
I tried a few... they are fairly cryptic and require good knowledge of biological terms...
- Bioinformatics Quick Start
- Making Sense of DNA and Protein Sequences
- Unmasking Genes in Human DNA
- Identification of Disease Genes
- Correlating Disease Genes and Phenoypes
- BLAST Quick Start
- EntrezGene Quick Start
- Structure Analysis Quick Start
- MapViewer Quick Start
- GenBank Quick Start
- Entrez Quick Start
- Microbial Genomes Quick Start
- Lab on Sequence Logo with Processing. Good reference can be found here.
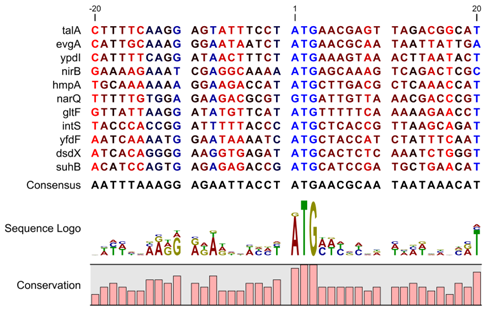
Image taken from http://www.clcbio.com/index.php?id=869