Difference between revisions of "CSC334 lab7"
| Line 7: | Line 7: | ||
[[Image:sequence_logo.png]] | [[Image:sequence_logo.png]] | ||
| + | <br /> | ||
| + | |||
This image is take from a the following document that you should read to get a good start on this lab: | This image is take from a the following document that you should read to get a good start on this lab: | ||
[http://www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/ www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/] | [http://www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/ www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/] | ||
| Line 33: | Line 35: | ||
</pre></code> | </pre></code> | ||
| − | ==First step: | + | ==First step: Skeleton Program and Window Geometry== |
More information will be provided during the lab. The goal of this step is to define the geometry of the window and the constants used by the program. | More information will be provided during the lab. The goal of this step is to define the geometry of the window and the constants used by the program. | ||
| − | + | <br /> | |
[[Image:lab7_window_geometry.png]] | [[Image:lab7_window_geometry.png]] | ||
| + | <br /> | ||
Create a new Processing sketchbook and paste in it the following skeleton program: | Create a new Processing sketchbook and paste in it the following skeleton program: | ||
| + | <br /> | ||
| + | |||
<code><pre> | <code><pre> | ||
// DNA_logo | // DNA_logo | ||
Revision as of 13:18, 4 August 2008
Contents
Introduction
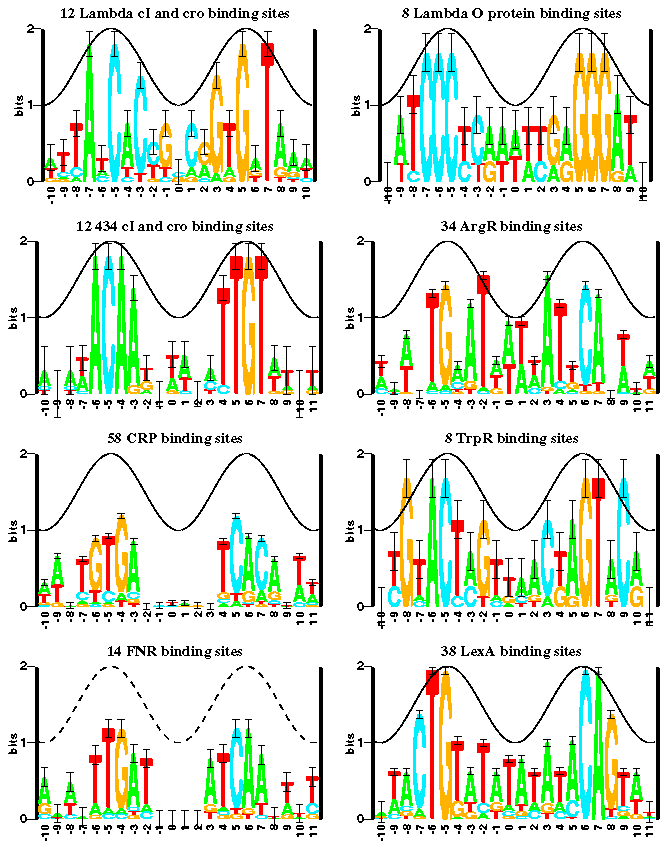
A good definition of sequence logos can be found in Wikipedia:
- A sequence logo in bioinformatics is a graphical representation of the sequence conservation of nucleotides (in a strand of DNA/RNA) or amino acids (in protein sequences) [1]
- To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. The sequence logo will show how well residues are conserved at each position: the fewer the number of residues, the higher the letters will be, because the better the conservation is at that position. Different residues at the same position will be scaled according to their frequency. Sequence logos can be used to represent conserved DNA binding sites, where transcription factors bind. [2]
This image is take from a the following document that you should read to get a good start on this lab: www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/
Lab
The Sequences
For this lab we will use 8 different sequences:
seq[0] = "CCCATTGTTCTC";
seq[1] = "TTTCTGGTTCTC";
seq[2] = "TCAATTGTTTAG";
seq[3] = "CTCATTGTTGTC";
seq[4] = "TCCATTGTTCTC";
seq[5] = "CCTATTGTTCTC";
seq[6] = "TCCATTGTTCGT";
seq[7] = "CCAATTGTTTTG";
They are shown here as taken from a Processing program where the sequences are stored in an array of 8 strings:
String seq[8];
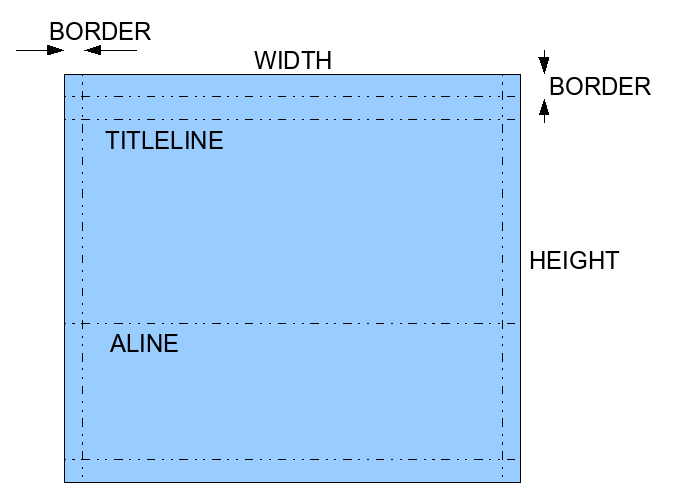
First step: Skeleton Program and Window Geometry
More information will be provided during the lab. The goal of this step is to define the geometry of the window and the constants used by the program.
Create a new Processing sketchbook and paste in it the following skeleton program:
// DNA_logo
// YourNameHere Date
//---------------------------------------------------------------------
// GEOMETRY
//---------------------------------------------------------------------
int WIDTH = ; // width of the window in pixels
int MIDWIDTH = WIDTH/2; // half that
int HEIGHT = ; // height, in pixels.
int BORDER = ; // border around the window where nothing
// is displayed
int TITLELINE = ; // y position of title line from top
int ALINE = ; // y position of line where logo appears
PFont font; // the font used to display the symbols
int NOSEQS = 8; // number of sequences
float Afreq[]; // frequency of A symbols in sequences
float Cfreq[]; // C
float Gfreq[]; // G
float Tfreq[]; // T
float information[]; // amount of information at each location
// of the consensus sequence
String seq[] = new String[NOSEQS]; // array of sequences
PImage a, c, g, t, black; // the 4 images for the 4 symbols
font = loadFont( "GillSans-60.vlw" ); // 60 points... very large!
textFont( font );
color myColor = color( 99, 66, 204 ); // font color
fill( myColor );
textSize( 24 ); // shrink for title
text( title, BORDER, TITLELINE ); // show title
}
//---------------------------------------------------------------------
// SETUP: called once when app starts.
//---------------------------------------------------------------------
void setup() {
size( WIDTH, HEIGHT );
background( 0, 0, 0 ); // black background
font = loadFont( "GillSans-24.vlw" ); // <== use your own font!
//--- load bitmap images for all 4 symbols ---
a = loadImage( "a.png" ); // load them from file into variables
c = loadImage( "c.png" );
g = loadImage( "g.png" );
t = loadImage( "t.png" );
//--- initialize all 8 sequences ---
seq[0] = "CCCATTGTTCTC";
seq[1] = "TTTCTGGTTCTC";
seq[2] = "TCAATTGTTTAG";
seq[3] = "CTCATTGTTGTC";
seq[4] = "TCCATTGTTCTC";
seq[5] = "CCTATTGTTCTC";
seq[6] = "TCCATTGTTCGT";
seq[7] = "CCAATTGTTTTG";
//--- generate arrays of frequencies and information ---
int noSymbols = seq[0].length( );
Afreq = new float[ noSymbols ];
Cfreq = new float[ noSymbols ];
Gfreq = new float[ noSymbols ];
Tfreq = new float[ noSymbols ];
information = new float[ noSymbols ];
//--- compute information at each position of sequence ---
findFreqsAndInformation();
//--- display the logo ---
displayLogo();
}
//---------------------------------------------------------------------
// displayLogo: displays the logo in the window, at y = ALINE.
//---------------------------------------------------------------------
void displayLogo( ) {
// ADD YOUR CODE HERE...
}
Pick good values for the constants.