CSC334 lab7
<meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> <meta name="description" content="DNA Sequence Logo Lab" /> <meta name="title" content="Bioinformatics Lab" /> <meta name="abstract" content="DNA Sequence Logo" /> <meta name="author" content="thiebaut at cs.smith.edu" />
Contents
- 1 Introduction
- 2 Lab
- 2.1 The Sequences
- 2.2 First step: Skeleton Program and Window Geometry
- 2.3 PNG images for the four symbols
- 2.4 The mechanics of printing a series of symbols using the PNG images
- 2.5 Displaying the correct symbols
- 2.6 Computing the frequency of each symbol in each position
- 2.7 Putting it all together
- 3 Solution Programs
Introduction
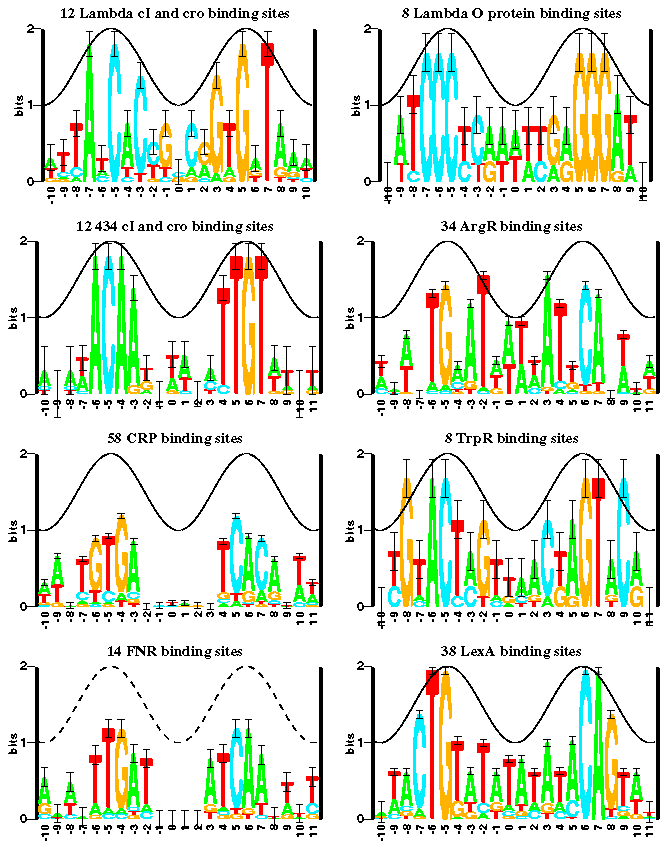
A good definition of sequence logos can be found in Wikipedia:
- A sequence logo in bioinformatics is a graphical representation of the sequence conservation of nucleotides (in a strand of DNA/RNA) or amino acids (in protein sequences) [1]
- To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. The sequence logo will show how well residues are conserved at each position: the fewer the number of residues, the higher the letters will be, because the better the conservation is at that position. Different residues at the same position will be scaled according to their frequency. Sequence logos can be used to represent conserved DNA binding sites, where transcription factors bind. [2]
This image is take from a the following document that you should read to get a good start on this lab: www-lmmb.ncifcrf.gov/~toms/how.to.read.sequence.logos/
Lab
The purpose of this lab is to create a Proce55ing sketchbook displaying the logo of a DNA sequence. Possible improvements include adding animation to the sketchbook to "grow" the logo, or add/remove sequences from the logo.
The Sequences
For this lab we will use 8 different sequences:
seq[0] = "CCCATTGTTCTC";
seq[1] = "TTTCTGGTTCTC";
seq[2] = "TCAATTGTTTAG";
seq[3] = "CTCATTGTTGTC";
seq[4] = "TCCATTGTTCTC";
seq[5] = "CCTATTGTTCTC";
seq[6] = "TCCATTGTTCGT";
seq[7] = "CCAATTGTTTTG";
They are shown here as taken from a Processing program where the sequences are stored in an array of 8 strings:
String seq[8];
First step: Skeleton Program and Window Geometry
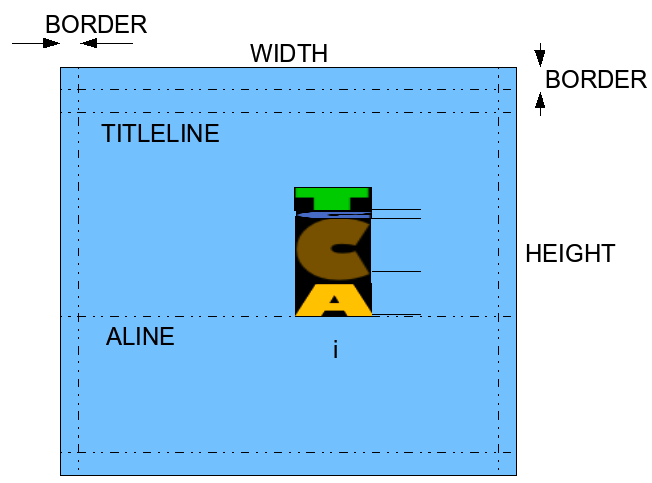
More information will be provided during the lab. The goal of this step is to define the geometry of the window and the constants used by the program.
Create a new Processing sketchbook and paste in it the following skeleton program:
// DNA_logo
// YourNameHere Date
//---------------------------------------------------------------------
// GEOMETRY
//---------------------------------------------------------------------
int WIDTH = ; // width of the window in pixels
int MIDWIDTH = WIDTH/2; // half that
int HEIGHT = ; // height, in pixels.
int BORDER = ; // border around the window where nothing
// is displayed
int TITLELINE = ; // y position of title line from top
int ALINE = ; // y position of line where logo appears
PFont font; // the font used to display the symbols
int NOSEQS = 8; // number of sequences
float Afreq[]; // frequency of A symbols in sequences
float Cfreq[]; // C
float Gfreq[]; // G
float Tfreq[]; // T
float information[]; // amount of information at each location
// of the consensus sequence
String seq[] = new String[NOSEQS]; // array of sequences
PImage a, c, g, t; // the 4 images for the 4 symbols
//---------------------------------------------------------------------
// SETUP: called once when app starts.
//---------------------------------------------------------------------
void setup() {
size( WIDTH, HEIGHT );
background( 0, 0, 0 ); // black background
font = loadFont( "GillSans-24.vlw" ); // <== use your own font!
textFont( font );
color myColor = color( 99, 66, 204 ); // font color
fill( myColor );
textSize( 24 );
text( "Did you remember to change this?", BORDER, TITLELINE ); // show title
//--- load bitmap images for all 4 symbols ---
a = loadImage( "a.png" ); // load them from file into variables
c = loadImage( "c.png" );
g = loadImage( "g.png" );
t = loadImage( "t.png" );
//--- initialize all 8 sequences ---
seq[0] = "CCCATTGTTCTC";
seq[1] = "TTTCTGGTTCTC";
seq[2] = "TCAATTGTTTAG";
seq[3] = "CTCATTGTTGTC";
seq[4] = "TCCATTGTTCTC";
seq[5] = "CCTATTGTTCTC";
seq[6] = "TCCATTGTTCGT";
seq[7] = "CCAATTGTTTTG";
//--- generate arrays of frequencies and information ---
int noSymbols = seq[0].length( );
Afreq = new float[ noSymbols ];
Cfreq = new float[ noSymbols ];
Gfreq = new float[ noSymbols ];
Tfreq = new float[ noSymbols ];
information = new float[ noSymbols ];
//--- display the logo ---
// ADD YOUR CODE HERE...
}
Pick good values for the constants. Run your program and fix possible syntax errors. Should anything display on the screen?) Also, make sure you create a font for your program using the Tools/Create Font utility.
PNG images for the four symbols
Processing does not have an easy way to print characters with a fixed width, but with a varying height. Processing, on the other hand, can easily shrink the height of an image. We will instead use four separate images representing each of the four symbols and draw them one of top of each other,
![]()
![]()
![]()
![]()
Get a copy of the 4 pictures and put them in the data folder of your Processing project.
The mechanics of printing a series of symbols using the PNG images
Assume that the string of symbols you want to display is
String sequence = "ACGTTTACCCTTAG"; int noSymbols = sequence.length();
Assuming that you want to display N symbols on the line we labeled ALINE in the geometry figure above, and that we want a margin equal to BORDER on each side of the line of symbols, how wide should the characters be?
int charWidth = ... ;
Let's set the height of the symbols to 60 for right now:
int charHeight = 60;
Displaying the symbols on the ALINE is done with a for-loop:
PImage img = a;
for ( int i=0; i< noSymbols; i++ )
image( img, BORDER + i*charWidth, ALINE, charWidth, charHeight );
Add this code at the end of the setup() function. Try it and verify that you get a line of A symbols.
Displaying the correct symbols
Use if-statements to display the correct symbol image. Comparing if the i-th symbol is an 'A' for example, can be done as follows:
if ( sequence.charAt( i )=='A' ) {
...
}
Modify your program and make it display all the symbols of the sequence.
Computing the frequency of each symbol in each position
If you look at the 8 sequences we have, you'll see that the symbol C appears 4 times in the first position. T appears also 4 times. A and G 0 times each. Their frequencies of occurrence is 0.5, 0.5, 0, and 0, respectively.
seq[0] = "CCCATTGTTCTC"; seq[1] = "TTTCTGGTTCTC"; seq[2] = "TCAATTGTTTAG"; seq[3] = "CTCATTGTTGTC"; seq[4] = "TCCATTGTTCTC"; seq[5] = "CCTATTGTTCTC"; seq[6] = "TCCATTGTTCGT"; seq[7] = "CCAATTGTTTTG";
Modify your program so that it computes the frequency of occurence of each symbol in each of the positions of the sequence. Record this frequency in the arrays of floats Afreq, Cfreq, Gfreq, and Tfreq.
You may check that your program is computing the correct information by making it print the contents of the arrays in the console window:
for ( int i=0; i< seq[0].length(); i++ ) {
println( "Afreq[" + i + "]=" + Afreq[i] );
}
The concensus sequence is the one made with the most likely symbol, given the different sequences. Given that we have four different symbols, we can define one symbol with only two bits. 'A' can be coded as 00, 'C' as 01, 'G' as 10, and 'T' as 11. If we are sure of the symbol that fits at position i in the consensus sequence, then the amount of information we have is 2 bits. If, as in the example above, the symbol is either 'C' or 'T', since they appear with equal frequency, then we know which symbol fits in the concensus sequence with only 1 bit of information.
The information in each position i is defined as: <math>Information( i ) = 2 + \sum_{sym=a,c,g,t} freq(sym) . \log_2( freq(sym) ) </math>
Refer to information given during the lab to compute this quantity for each symbol.
Make your program print the information found for each consensus position in the console window, as you did for the frequencies of the symbols.
Putting it all together
Note 1: Normally the logo always displays the tallest symbol on the top of the stack of symbols. The program shown here always displays the symbol in the same order, irrespective of their respective heights.
Display the symbols for the consensus sequence in such a way that all four symbols are displayed one on top of each other, and such that their height is proportional to their frequency and the amount of information at that position.
More information will be provided during the lab. Use the following picture as reference:
And if it all works well...