CSC334 Lab10
<meta name="keywords" content="computer science, bioinformatics, DNA, CSC334, Lab" /> <meta name="description" content="DNA Sequence Logo Lab" /> <meta name="title" content="Bioinformatics Lab" /> <meta name="abstract" content="DNA Sequence Logo" /> <meta name="author" content="thiebaut at cs.smith.edu" />
Finding the Secondary Structure of a Protein
The page on Secondary Structures in Wikipedia defines it well:
- In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids (DNA/RNA). It does not, however, describe specific atomic positions in three-dimensional space, which are considered to be tertiary structure.
- Secondary structure is formally defined by the hydrogen bonds of the biopolymer, as observed in an atomic-resolution structure. In proteins, the secondary structure is defined by patterns of hydrogen bonds between backbone amide and carboxyl groups (sidechain-mainchain and sidechain-sidechain hydrogen bonds are irrelevant), where the DSSP definition of a hydrogen bond is used. In nucleic acids, the secondary structure is defined by the hydrogen bonding between the nitrogenous bases.
Goal
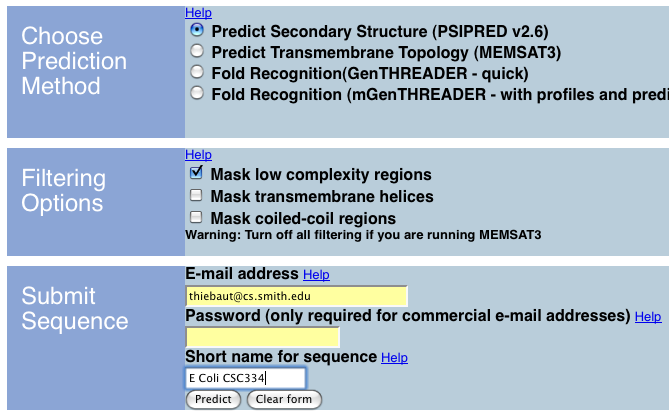
In this lab we use the PSIPRED system to get the secondary structure of a protein.
Steps
- Connect to the Psipred system at bioinf.cs.ucl.ac.uk/psipred/
- Click on CLICK HERE TO ENTER THE SERVER
- Copy/Paste a protein sequence in FASTA format (without the first text line). Here we are using a sequence we obtained in Lab #9 for the E. Coli sequence reference AAF33488:
MESTVDTELLKTFLEVSRTRHFGRAAEALYLTQSAVSFRIRQLENQLGVNLFTRHRNNIRLTTAGEKLLP YAETLMNTWQAARKEVAHTSRHNEFSIGASASLWECMLNAWLGRLYQLQEPQSGLQFEARIAQRQSLVKQ LHERQLDLLITTEAPKMDEFSSQLLGHFTLALYCSSPARKKSELNYLRLEWGPDFQQHETGLIAADEVPV LTTSSAELARQQLSALNGCSWLPVNWANEKGGLHTVADSATLSRPLYAIWLQNSDKYSLICDLLKTDVLD EQ
- Accept the defaults, enter your email address, then click on Predict
- Wait a while
- Wait some more
- Depending on the number of job requests at Psipred, you may have to wait quite a while. If this is too long and you want to continue with one of the sequences I was able to get processed ahead of time and get back via email, follow this link.
- Assuming that you have received the result back from Psipred, you will observe that you get several pieces of information, including text, pdf, and jpeg outputs.
- Observe the text output. It should look something like this:
PSIPRED PREDICTION RESULTS
Key
Conf: Confidence (0=low, 9=high)
Pred: Predicted secondary structure (H=helix, E=strand, C=coil)
AA: Target sequence
# PSIPRED HFORMAT (PSIPRED V2.6 by David Jones)
Conf: 999888999999999998399899999967888489999999999829843897699436
Pred: CCCCCCHHHHHHHHHHHHCCCHHHHHHHHCCCCCHHHHHHHHHHHHHCCEEEEECCCCEE
AA: MESTVDTELLKTFLEVSRTRHFGRAAEALYLTQSAVSFRIRQLENQLGVNLFTRHRNNIR
10 20 30 40 50 60
. . .
Conf: 988943789986148999995899707999999998764059
Pred: CCEEECCCCCCCEEEEEEEEECCCCCHHHHHHHHHHHHHHCC
AA: GGLHTVADSATLSRPLYAIWLQNSDKYSLICDLLKTDVLDEQ
250 260 270 280
- Question 1: Print your query result on a piece of paper and color the helix, strand, and coil regions of the protein with different colors.
- Question 2: You will have noticed that the first line of each group is a confidence value.
For the secondary structure that you got back for your query, what is the overal, average confidence value for the prediction of regions to helix, coil or strand structure? Be precise, and figure out how to use the computer to give you the answer!
- Question 3: Display/print the pdf or jpeg versions you will have received from Psipred.