Difference between revisions of "CSC352 Homework 5 2013"
| (29 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
---- | ---- | ||
| − | <bluebox>This assignment is due on 11/14 at 11:59 p.m.</bluebox> | + | <bluebox>This assignment is due on 11/14 at 11:59 p.m. Its purpose is to write an MPI program on Amazon's AWS system to identify a large collection of images. This assignment skips the storing of the geometry in a MySQL database.</bluebox> |
<br /> | <br /> | ||
=Assignment= | =Assignment= | ||
<br /> | <br /> | ||
| − | This assignment is in two parts. They are both identical, but you have to do both | + | This assignment is in two parts. They are both identical, but you have to do both: |
| − | + | * The first part is to develop an MPI program on Hadoop0. | |
| − | The first part is to develop an MPI program on Hadoop0. | + | * The second part is to port this program to AWS. |
| − | |||
| − | The second part is to port this program to AWS. | ||
It is important to debug programs on local machines rather than AWS otherwise the money is spend on development time rather than production time. For us, for this assignment, the ''production'' is to calculate the geometry of a large set of images. | It is important to debug programs on local machines rather than AWS otherwise the money is spend on development time rather than production time. For us, for this assignment, the ''production'' is to calculate the geometry of a large set of images. | ||
<br /> | <br /> | ||
| + | |||
=On Hadoop0= | =On Hadoop0= | ||
<br /> | <br /> | ||
| Line 25: | Line 24: | ||
* Remove the part of the program that stores the geometry in the database. Your program will have the manager send block of file names to the workers, the workers will use ''identify'' to get the geometry of each file, and will simply drop that information and not store it anywhere. That's okay for this assignment. | * Remove the part of the program that stores the geometry in the database. Your program will have the manager send block of file names to the workers, the workers will use ''identify'' to get the geometry of each file, and will simply drop that information and not store it anywhere. That's okay for this assignment. | ||
| − | * Set the number of nodes to 8: 1 manager and 7 workers. | + | * Set the number of nodes to 8: 1 manager and 7 workers. This should work well on the 8-core Hadoop0 processor. |
* Figure out how many images ''N'' to process with 8 nodes so that the computation of their geometry takes less than a minute, but more than 10 seconds. | * Figure out how many images ''N'' to process with 8 nodes so that the computation of their geometry takes less than a minute, but more than 10 seconds. | ||
| − | * Run a series of experiments to figure out the size of the packet of file names exchanged between the manager and a worker that yields the | + | * Run a series of experiments to figure out the size of the packet of file names exchanged between the manager and a worker that yields the fastest real execution-time. In other words, run the MPI program on hadoop0 and pass it a value of ''M'' equal to, say, 10. This means that the manager will walk the directories of images and grab 10 consecutive images and pass (MPI_Send) their names in one packet to a worker. It will then grab another 10 images and pass that packet to another worker. etc. Measure the time it takes your program to process ''N'' images. Repeat the same experiment for other values of ''M'' ranging from 10 to 5,000. For example 10, 50, 100, 250, 500, 1000, 2500, 5000. |
| − | ''M'' ranging from 10 to 5,000. For example 10, 50, 100, 250, 500, 1000, 2500, 5000. | ||
Once your program runs correctly on hadoop0, port it to an MPI cluster on AWS that you will start with the '''starcluster''' utility. | Once your program runs correctly on hadoop0, port it to an MPI cluster on AWS that you will start with the '''starcluster''' utility. | ||
<br /> | <br /> | ||
| − | |||
<br /> | <br /> | ||
| − | |||
| − | == | + | =On the AWS Cluster= |
| + | <br /> | ||
| + | You will need to modify the program and the starcluster config file to fully port your program to AWS. The setup we want to implement is illustrated in the figure below: | ||
| + | <br /> | ||
| + | <center>[[Image:ClusterWith2EBSVolumes.jpg|600px]]</center> | ||
| + | <br /> | ||
| + | ==EBS Volume== | ||
| + | <br /> | ||
| + | You should create a 1-GByte EBS for yourself, where you can keep your program files. Files stored in the default directories of the cluster will disappear when you terminate the cluster. To keep files around on AWS, you need to store them in Elastic Block Devices, or '''EBS''' volumes. | ||
| + | Follow the directions (slightly modified since we did the lab on AWS) from [[Create_an_MPI_Cluster_on_the_Amazon_Elastic_Cloud_(EC2)#Creating_an_EBS_Volume| this section]] and the [[Create_an_MPI_Cluster_on_the_Amazon_Elastic_Cloud_(EC2)#Attaching_the_EBS_Volume_to_the_Cluster| the section that follows]] to attach your personal '''data''' EBS volume as well as the '''enwiki''' volume to your cluster. | ||
| + | <br /> | ||
| + | ==Cluster Size== | ||
| + | <br /> | ||
Modify the config file in the ~/.starcluster directory on your mac and set the number of nodes to 8. The instance type should be m1.medium. | Modify the config file in the ~/.starcluster directory on your mac and set the number of nodes to 8. The instance type should be m1.medium. | ||
| + | <br /> | ||
| + | ==Config File== | ||
| + | <br /> | ||
| + | Your config file should look something like this (I have removed all unnecessary comments), and anonymized personal information. The areas in magenta are the places you will need to enter your own data or data provided here (for example the enwiki volume is fixed). | ||
| + | |||
| + | <onlysmith> | ||
| + | #################################### | ||
| + | ## StarCluster Configuration File ## | ||
| + | #################################### | ||
| + | [global] | ||
| + | DEFAULT_TEMPLATE=smallcluster | ||
| + | |||
| + | ############################################# | ||
| + | ## AWS Credentials and Connection Settings ## | ||
| + | ############################################# | ||
| + | [aws info] | ||
| + | AWS_ACCESS_KEY_ID = <font color="magenta">XXXXXXXXXXXXXXXXXXXXXX </font> | ||
| + | AWS_SECRET_ACCESS_KEY = <font color="magenta">abcdefghijklmnopqrstuvwxyzABCDEFGHIJKLMNOPQRSTUVWXYZ</font> | ||
| + | AWS_USER_ID= <font color="magenta">mickeymouse</font> | ||
| + | |||
| + | ########################### | ||
| + | ## Defining EC2 Keypairs ## | ||
| + | ########################### | ||
| + | [key <font color="magenta">mykeyABC</font>] | ||
| + | KEY_LOCATION=<font color="magenta">~/.ssh/mykeyABC.rsa</font> | ||
| + | |||
| + | ################################ | ||
| + | ## Defining Cluster Templates ## | ||
| + | ################################ | ||
| + | [cluster smallcluster] | ||
| + | KEYNAME = <font color="magenta">mykeyABC</font> | ||
| + | CLUSTER_SIZE = <font color="magenta">8</font> | ||
| + | CLUSTER_USER = sgeadmin | ||
| + | CLUSTER_SHELL = bash | ||
| + | NODE_IMAGE_ID = ami-7c5c3915 | ||
| + | NODE_INSTANCE_TYPE = m1.medium | ||
| + | VOLUMES = <font color="magenta">dataABC</font>, enwiki | ||
| + | #SPOT_BID = 0.50 | ||
| + | |||
| + | ############################# | ||
| + | ## Configuring EBS Volumes ## | ||
| + | ############################# | ||
| + | [volume enwiki] | ||
| + | VOLUME_ID = <font color="magenta">vol-XXXXXX </font> #*** SEE NOTE BELOW *** | ||
| + | MOUNT_PATH = /enwiki/ | ||
| + | |||
| + | [volume <font color="magenta">dataABC</font>] | ||
| + | VOLUME_ID = <font color="magenta">vol-eeeeee</font> | ||
| + | MOUNT_PATH = /data/ | ||
| + | |||
| − | |||
<br /> | <br /> | ||
| − | + | <tanbox> | |
| + | ;'''Copy of Note posted to PIAZZA on 11/12/13 @ 9:40 p.m.:'''<br /> | ||
| + | :We found out on 11/12/13 that we couldn't share the same /enwiki mounted EBS containing the image files. This is the /enwiki directory on AWS introduced in this homework. | ||
| − | + | :So, I have created 6 new EBS volumes with a copy of the image files. You will each have your own EBS volume mounted on the /enwiki directory on your MPI cluster. | |
| − | + | :Below is the section that defines what volume is mounted on the /enwiki directory of your cluster: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[volume enwiki] | [volume enwiki] | ||
| − | VOLUME_ID = vol- | + | VOLUME_ID = vol-xxxxxxxx |
MOUNT_PATH = /enwiki | MOUNT_PATH = /enwiki | ||
| − | + | ||
| − | + | :You just need to change the vol-xxxxx with the id shown below: | |
| − | + | ||
| − | + | Danae's volume: '''vol-3ded877e''' | |
| − | + | Emily's volume: '''vol-46e38905''' | |
| + | Gavi's volume: '''vol-30e28873''' | ||
| + | Julia's volume: '''vol-8bec86c8''' | ||
| + | Sharon Pamela's volume: '''vol-b1e288f2''' | ||
| + | Yoshie's volume: '''vol-02e28841''' | ||
| + | |||
| + | </tanbox> | ||
</onlysmith> | </onlysmith> | ||
| + | <br /> | ||
| − | + | ==Images== | |
| + | <br /> | ||
| + | A sample (150,000 or so) of the 3 million images have been transferred to the '''enwiki EBS drive''' in our AWS environment. Once you have edited your configuration file as shown above with the '''enwiki''' volume, the images files should be available to your program in the /enwiki directory on the cluster. | ||
| + | |||
| + | Next, start the cluster; you should have two directories that will appear in the root directory, '''/data''', and '''/enwiki'''. All nodes will have access to both of them. Approximately 150,000 images have already been uploaded to the directory '''/enwiki/''', in three subdirectories, '''0''', '''1''', and '''2'''. | ||
To get a sense of where the images are, start your cluster with just 1 node (no need to create a large cluster just to explore the system), and ssh to the master: | To get a sense of where the images are, start your cluster with just 1 node (no need to create a large cluster just to explore the system), and ssh to the master: | ||
| Line 79: | Line 143: | ||
<br /> | <br /> | ||
| + | |||
=ImageMagick and Identify= | =ImageMagick and Identify= | ||
<br /> | <br /> | ||
| Line 90: | Line 155: | ||
And identify will be installed on the master. Unfortunately, you'll have to install it as well on all the workers. If you '''stop''' your cluster and not '''terminate''' it, the installation will remain until the next time you restart your cluster. If you '''terminate''' your cluster, however, you'll have to reinstall imagemagick the next time to start your cluster. | And identify will be installed on the master. Unfortunately, you'll have to install it as well on all the workers. If you '''stop''' your cluster and not '''terminate''' it, the installation will remain until the next time you restart your cluster. If you '''terminate''' your cluster, however, you'll have to reinstall imagemagick the next time to start your cluster. | ||
| + | <br /> | ||
<br /> | <br /> | ||
=Measurements= | =Measurements= | ||
<br /> | <br /> | ||
| − | + | Perform the same measurements you did on Hadoop0 and measure the execution times (real time) for different values of ''M''. | |
| − | + | You may have to pick a different value of ''N'' that will make the execution time not exceed a minute. | |
| − | + | Plot the execution times as a function of M, and submit a jpg/png/pdf version of it via email. Use '''CSC352 Homework 5 graph''' for subject of your email message, please! | |
| + | <br /> | ||
| + | =Submission= | ||
| + | <br /> | ||
| + | Call your program hw5aws.c and submit it from your 352a-xx account on beowulf as follows: | ||
| − | <br /><br /><br /><br /><br /> | + | submit hw5 hw5aws.c |
| + | |||
| + | If you created additional files (such as shell files), submit them as well. | ||
| + | <br /> | ||
| + | =Optional and Extra Credit= | ||
| + | <br /> | ||
| + | The program we saw in class uses a ''round-robin'' approach to feed data to the workers. Modify the program so that it will send packets of file names to a worker that is idle, rather than to the next logical one. | ||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | <tanbox>ALWAYS REMEMBER TO STOP YOUR CLUSTER WHEN YOU ARE NOT USING IT!</tanbox> | ||
| + | |||
| + | <br /> | ||
=Misc. Information= | =Misc. Information= | ||
| − | + | <br /> | |
In case you wanted to have the MPI program store the image geometry in your database, you'd have to follow the process described in [[Tutorial:_C_%2B_MySQL_%2B_MPI#Verify_that_the_MySQL_API_works| this tutorial]]. However, if you were to create the program '''mysqlTest.c''' on your AWS cluster, you'd find that the command '''mysql_config''' is not installed on the default AMI used by starcluster to create the MPI cluster. | In case you wanted to have the MPI program store the image geometry in your database, you'd have to follow the process described in [[Tutorial:_C_%2B_MySQL_%2B_MPI#Verify_that_the_MySQL_API_works| this tutorial]]. However, if you were to create the program '''mysqlTest.c''' on your AWS cluster, you'd find that the command '''mysql_config''' is not installed on the default AMI used by starcluster to create the MPI cluster. | ||
| Line 121: | Line 203: | ||
images2 | images2 | ||
pics1 | pics1 | ||
| + | |||
| + | <br /> | ||
| + | =Extending Homework to Project= | ||
| + | <br /> | ||
| + | This homework assignment can be the beginning of a project. Just one option for you to consider. The description below could very well apply to some other part of our collage project. | ||
| + | |||
| + | There would be several parts needed to expand the current homework to a project: | ||
| + | * You could continue the extra-credit part, and implement a ''double-buffer'' system for the communication between the manager and its workers. In this approach the manager sends two data blocks to a worker to start it going. When the worker is done with one block, it indicates this status to the manager which sends another block while the worker continues processing the second block. For this to work well you need to use non blocking variants of the MPI_Send and MPI_Recv primitives. | ||
| + | * You would need to figure out which database to use. You could either use the one you own on Hadoop0, or you could figure out how to create one on the AWS and store it on an EBS volume. | ||
| + | * You would need to tune the system by running several experiments that would indicate a range of performance-optimizing values for one of the parameters controlling your system. Such parameters could be: | ||
| + | ** The size of the block (i.e. the number of file names sent by the manager to a worker in a given block) | ||
| + | ** The size of the cluster (number of nodes) | ||
| + | ** The type of instance used versus price. You can bet that using a cluster of 10 cr1.8xlarge instances with an equivalent 88 ECUs each would work faster than a cluster of 10 m1.medium instances with an equivalent of 2 ECUs each. But 10 cr1.8xlarge instances will cost $35 an hour, while 10 m1.medium instances will cost you $1.20 an hour. If you're willing to spend $35 an hour, then you could actually create a cluster of 291 m1.medium instances. In this case, what cluster would allow you to identify the largest number of images in one hour, given a fixed cost? | ||
| + | <br /><br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | [[Category:CSC352]][[Category:Homework]][[Category:MPI]] | ||
| + | <onlydft> | ||
| + | To see which file takes the longest to be processed by identify: | ||
| + | |||
| + | for i in `ls *`; do | ||
| + | x=$( /usr/bin/time -f "%e" identify -quiet $i 2>&1 | tail -1 ) | ||
| + | echo $x $i | ||
| + | done | sort -n | tail | ||
| + | 0.06 Pangil_bamboo_ferryman.jpg | ||
| + | 0.07 Central_College_(Iowa)_logo.svg | ||
| + | 0.07 Survivor-baby-wallaby.jpg | ||
| + | 0.07 Westlake_High_School_emblem.svg | ||
| + | 0.08 ChokerSetters.jpg | ||
| + | 0.08 Pakistan_France_Locator.svg | ||
| + | 0.08 Samoa_USA_Locator.svg | ||
| + | 0.08 SE-Scale_full.jpg | ||
| + | 0.10 Mexico_344.jpg | ||
| + | 0.26 House_Resoultion_167.pdf | ||
| + | |||
| + | </onlydft> | ||
Latest revision as of 23:14, 14 November 2013
--D. Thiebaut (talk) 20:06, 4 November 2013 (EST)
Contents
Assignment
This assignment is in two parts. They are both identical, but you have to do both:
- The first part is to develop an MPI program on Hadoop0.
- The second part is to port this program to AWS.
It is important to debug programs on local machines rather than AWS otherwise the money is spend on development time rather than production time. For us, for this assignment, the production is to calculate the geometry of a large set of images.
On Hadoop0
- Take the MPI program we studied in class, and available here, and modify it so that it takes two parameters from the command line:
- the total number of images to process, N, and
- the number M of image names sent by the manager to its workers in one packet.
- Note that on hadoop0 the program expects the images to be stored in the directory /media/dominique/3TB/mediawiki/images/wikipedia/en
- Remove the part of the program that stores the geometry in the database. Your program will have the manager send block of file names to the workers, the workers will use identify to get the geometry of each file, and will simply drop that information and not store it anywhere. That's okay for this assignment.
- Set the number of nodes to 8: 1 manager and 7 workers. This should work well on the 8-core Hadoop0 processor.
- Figure out how many images N to process with 8 nodes so that the computation of their geometry takes less than a minute, but more than 10 seconds.
- Run a series of experiments to figure out the size of the packet of file names exchanged between the manager and a worker that yields the fastest real execution-time. In other words, run the MPI program on hadoop0 and pass it a value of M equal to, say, 10. This means that the manager will walk the directories of images and grab 10 consecutive images and pass (MPI_Send) their names in one packet to a worker. It will then grab another 10 images and pass that packet to another worker. etc. Measure the time it takes your program to process N images. Repeat the same experiment for other values of M ranging from 10 to 5,000. For example 10, 50, 100, 250, 500, 1000, 2500, 5000.
Once your program runs correctly on hadoop0, port it to an MPI cluster on AWS that you will start with the starcluster utility.
On the AWS Cluster
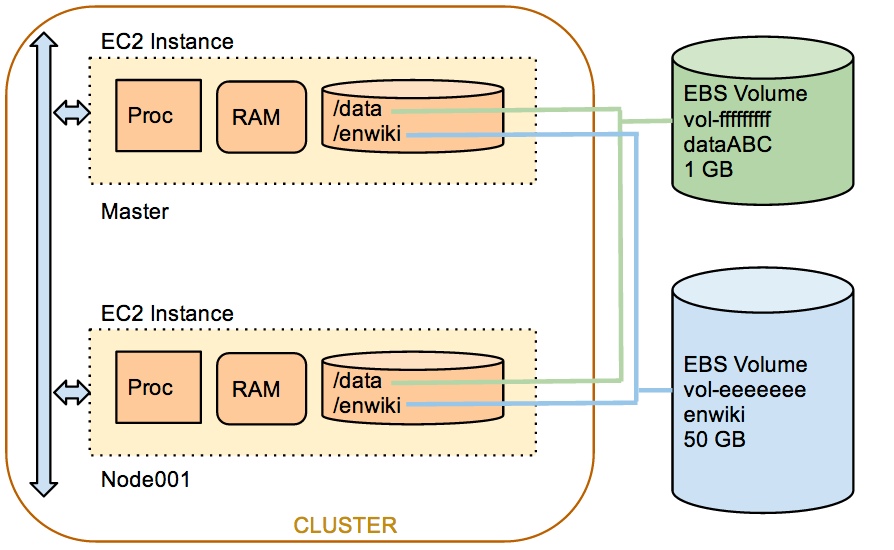
You will need to modify the program and the starcluster config file to fully port your program to AWS. The setup we want to implement is illustrated in the figure below:
EBS Volume
You should create a 1-GByte EBS for yourself, where you can keep your program files. Files stored in the default directories of the cluster will disappear when you terminate the cluster. To keep files around on AWS, you need to store them in Elastic Block Devices, or EBS volumes.
Follow the directions (slightly modified since we did the lab on AWS) from this section and the the section that follows to attach your personal data EBS volume as well as the enwiki volume to your cluster.
Cluster Size
Modify the config file in the ~/.starcluster directory on your mac and set the number of nodes to 8. The instance type should be m1.medium.
Config File
Your config file should look something like this (I have removed all unnecessary comments), and anonymized personal information. The areas in magenta are the places you will need to enter your own data or data provided here (for example the enwiki volume is fixed).
Images
A sample (150,000 or so) of the 3 million images have been transferred to the enwiki EBS drive in our AWS environment. Once you have edited your configuration file as shown above with the enwiki volume, the images files should be available to your program in the /enwiki directory on the cluster.
Next, start the cluster; you should have two directories that will appear in the root directory, /data, and /enwiki. All nodes will have access to both of them. Approximately 150,000 images have already been uploaded to the directory /enwiki/, in three subdirectories, 0, 1, and 2.
To get a sense of where the images are, start your cluster with just 1 node (no need to create a large cluster just to explore the system), and ssh to the master:
starcluster start mycluster starcluster sshmaster mycluster ls /enwiki ls /enwiki/0 ls /enwiki/0/01 etc...
ImageMagick and Identify
Identify is a utility that is part of Imagemagick. Unfortunately, Imagemagick is not installed by default on our AWS clusters. Doing image processing is apparently something not regularly performed by mpi programs. But installing it is easy:
On the master node, type
apt-get update apt-get install imagemagick
And identify will be installed on the master. Unfortunately, you'll have to install it as well on all the workers. If you stop your cluster and not terminate it, the installation will remain until the next time you restart your cluster. If you terminate your cluster, however, you'll have to reinstall imagemagick the next time to start your cluster.
Measurements
Perform the same measurements you did on Hadoop0 and measure the execution times (real time) for different values of M.
You may have to pick a different value of N that will make the execution time not exceed a minute.
Plot the execution times as a function of M, and submit a jpg/png/pdf version of it via email. Use CSC352 Homework 5 graph for subject of your email message, please!
Submission
Call your program hw5aws.c and submit it from your 352a-xx account on beowulf as follows:
submit hw5 hw5aws.c
If you created additional files (such as shell files), submit them as well.
Optional and Extra Credit
The program we saw in class uses a round-robin approach to feed data to the workers. Modify the program so that it will send packets of file names to a worker that is idle, rather than to the next logical one.
Misc. Information
In case you wanted to have the MPI program store the image geometry in your database, you'd have to follow the process described in this tutorial. However, if you were to create the program mysqlTest.c on your AWS cluster, you'd find that the command mysql_config is not installed on the default AMI used by starcluster to create the MPI cluster.
To install the mysql_config utility, run the following commands on the master node of your cluster as root:
apt-get update apt-get build-dep python-mysqldb
Edit the constants in mysqlTest.c that define the address of the database server (hadoop0), as well as the credentials of your account on the mysql server.
You can then compile and run program:
gcc -o mysqlTest $(mysql_config --cflags) mysqlTest.c $(mysql_config --libs) ./mysqlTest MySQL Tables in mysql database: images images2 pics1
Extending Homework to Project
This homework assignment can be the beginning of a project. Just one option for you to consider. The description below could very well apply to some other part of our collage project.
There would be several parts needed to expand the current homework to a project:
- You could continue the extra-credit part, and implement a double-buffer system for the communication between the manager and its workers. In this approach the manager sends two data blocks to a worker to start it going. When the worker is done with one block, it indicates this status to the manager which sends another block while the worker continues processing the second block. For this to work well you need to use non blocking variants of the MPI_Send and MPI_Recv primitives.
- You would need to figure out which database to use. You could either use the one you own on Hadoop0, or you could figure out how to create one on the AWS and store it on an EBS volume.
- You would need to tune the system by running several experiments that would indicate a range of performance-optimizing values for one of the parameters controlling your system. Such parameters could be:
- The size of the block (i.e. the number of file names sent by the manager to a worker in a given block)
- The size of the cluster (number of nodes)
- The type of instance used versus price. You can bet that using a cluster of 10 cr1.8xlarge instances with an equivalent 88 ECUs each would work faster than a cluster of 10 m1.medium instances with an equivalent of 2 ECUs each. But 10 cr1.8xlarge instances will cost $35 an hour, while 10 m1.medium instances will cost you $1.20 an hour. If you're willing to spend $35 an hour, then you could actually create a cluster of 291 m1.medium instances. In this case, what cluster would allow you to identify the largest number of images in one hour, given a fixed cost?